| PATHOME-Drug overview |
|
- Drug repositioning aims at revealing novel indications for existing drugs, in particular, in case of diseases with no available drugs. Diverse computational drug repositioning methods have been proposed by measuring either drug-treated gene expression signatures or proximity of drug targets and disease proteins in prior networks. However, these methods do not explain which signaling subparts allow potential drugs to be selected, and not consider polypharmacology, i.e., multiple targets of a known drug, in the specific subparts.
- PATHOME-Drug locates subparts of signaling cascading relating to phenotype changes (e.g., disease status changes), and obtains existing drugs such that their multiple targets are enriched in the subparts. Our method demonstrated better performances for signaling context detection and drug/compound detection, in comparison with a drug repositioning web service, WebGestalt, for both real biological and simulated datasets. We believe that our tool can be recognized in the current situation of shortage of targeted therapy agents.
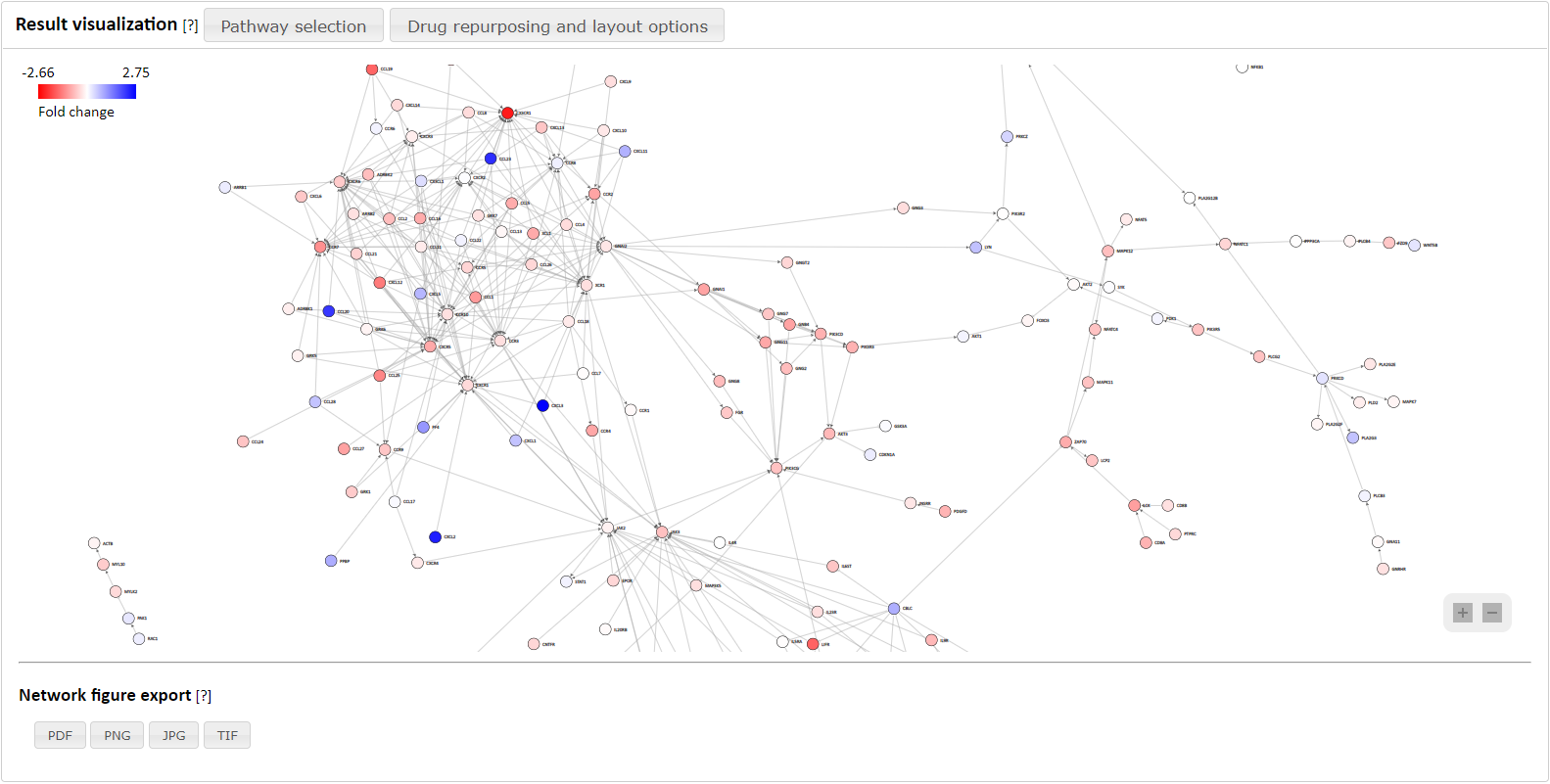 Figure 1.
Figure 1. Extraction of network from big data repository and its visualization.
Note: This section contains some parts of our previous publication (Oncogene (2014), 33, 4941-4951) under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License.
|
|