HisCoM-PAGE
BIBS lab, Seoul National University

Main Page Usage Tutorial Download
Usage
HisCoM-PAGE is written by R and can be installed by the following steps.
Step 1. Source Code Download
HisCoM-PAGE can be run by R program. To run the HisCoM-PAGE, you should first download theDownload zipfile which contains ‘program code.R’, data_preparation.R, and sample data. The ‘program code.R’ contains following four functions.
- CV_HisCoM ------------------------------------------------- Implements HisCoM with cross-validation.
- Logliklihood_calculation ----------------------- Partial log-likelihood calculation for survival model.
- Estimation_code ---------------------------------------- Estimates the pathway and gene coefficient.
- HisCoM _PAGE ----------------------------------------------------------------------------------Main function.
Step 2. Data File Preparation
Step 2 is to import the data files of pathway information, gene expression data, and trait file.
Pathway database file
Users should prepare the pathway database file using pathway database. An example of pathway database file is given below. The first column shows the name of pathways and the other columns the gene names belong to the pathways.

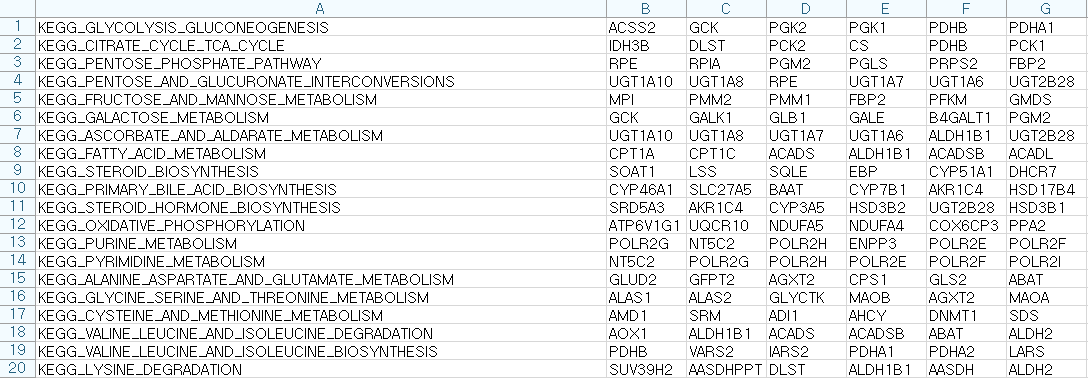
Fig1. Pathway Database example
(Source : http://software.broadinstitute.org/gsea/msigdb/collections.jsp#C2)
Gene expression data
Users should prepare the gene expression data (Filtering step is optional). Gene expression data can be either RNAseq data or microarray data. The first column represents genes and the other columns samples.
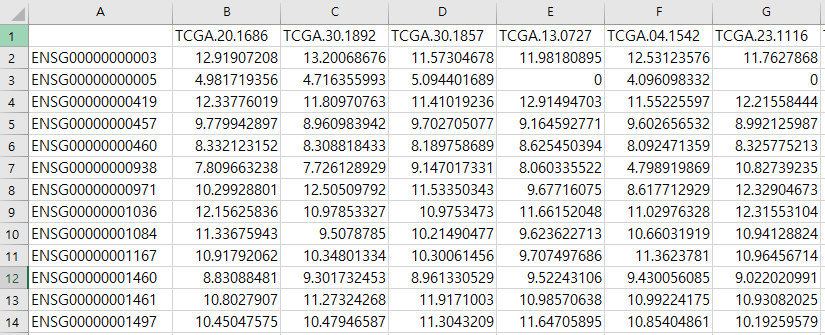
Fig2. Gene expression data example (Source https://portal.gdc.cancer.gov)
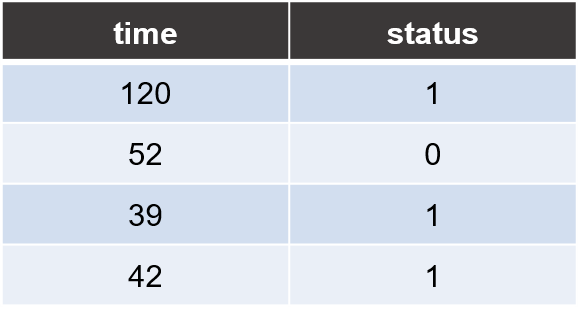
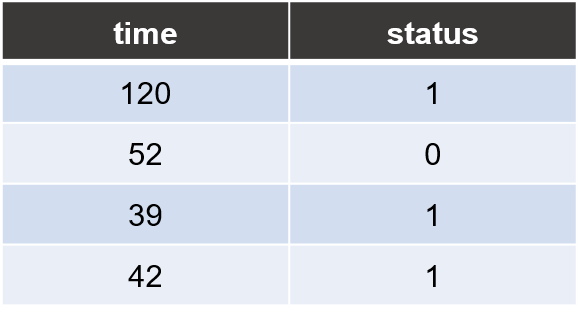
Trait file
Each line consists of two columns for sample’s survival time and censoring status, respectively.
Step 3. Input File for HisCoM-PAGE
Using three files from Step 2, users should prepare three input files for running HisCoM-PAGE. Three input files are required to run HisCoM-PAGE. The following table shows the summary of three files.
|
pathway_annot file (csv,RDS etc.) |
Contains (1) pathway name and (2) gene name. You should make the same format as example pathway annot file. |
|
gene_expression file (csv,RDS etc.) |
Contains normalized gene expression values. The format is same with the ‘Input file- Gene expression file.’ |
|
trait file (csv,RDS etc.) |
Contains (1) survival time and (2) censoring status. |
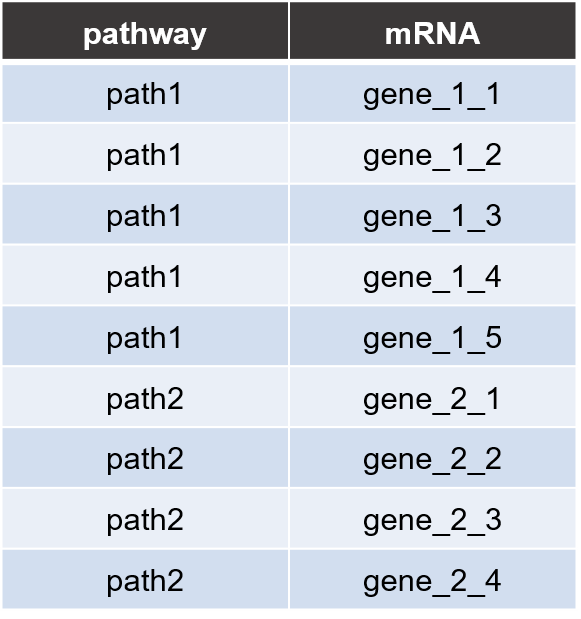
Pathway annot file
Pathway database parsing example code is given in ‘data_preparation.R’ program. Using the parsing code, users can make pathway annot file. Each line consists of two columns for pathway name and gene name, respectively.
Trait file
Each line consists of two columns for sample’s survival time and censoring status, respectively.
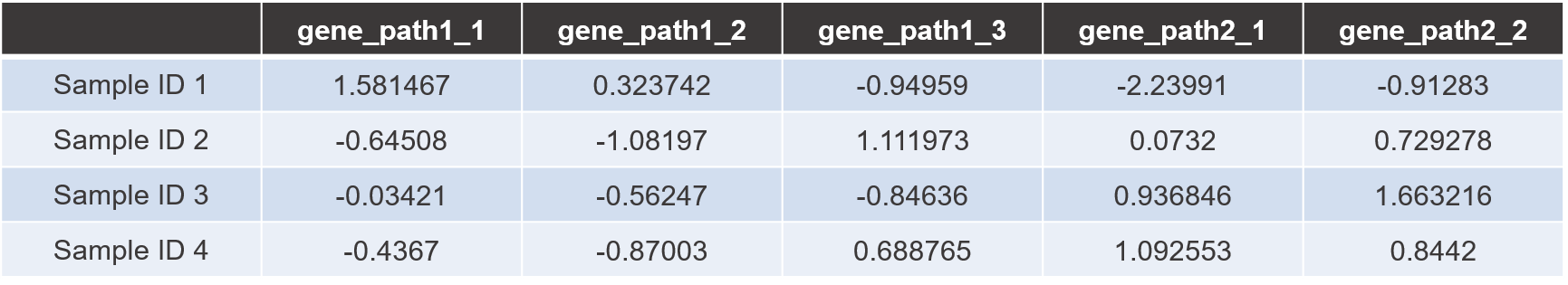
Gene expression file
Data parsing example code is given in ‘data_preparation.R’ program file. Using the parsing code, users can make ‘gene expression input file’ for HisCoM-PAGE.
Row represents the sample name and the column represents the pathway matched gene name and each cell represents the normalized gene expression value.
Step 4. Results
Using three files from Step 4, HisCoM-PAGE can be run. The followings are the output.
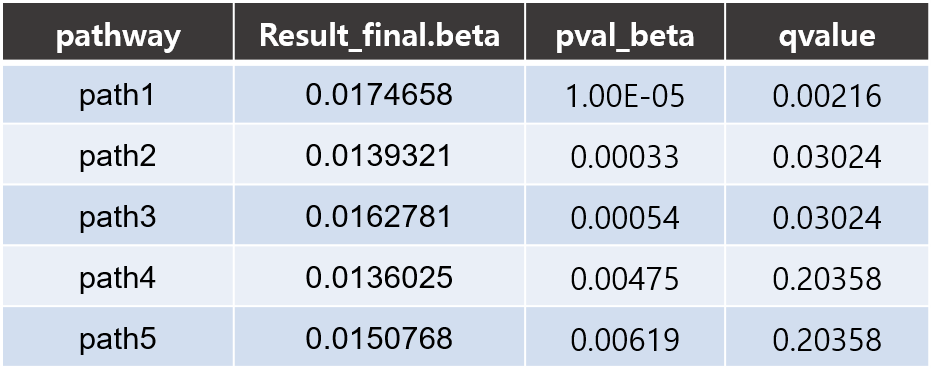
- Pathway results
- csv: Each line consists of four columns for pathway name, pathway coefficient (β_path), permutation p-value and FDR corrected q-value respectively.
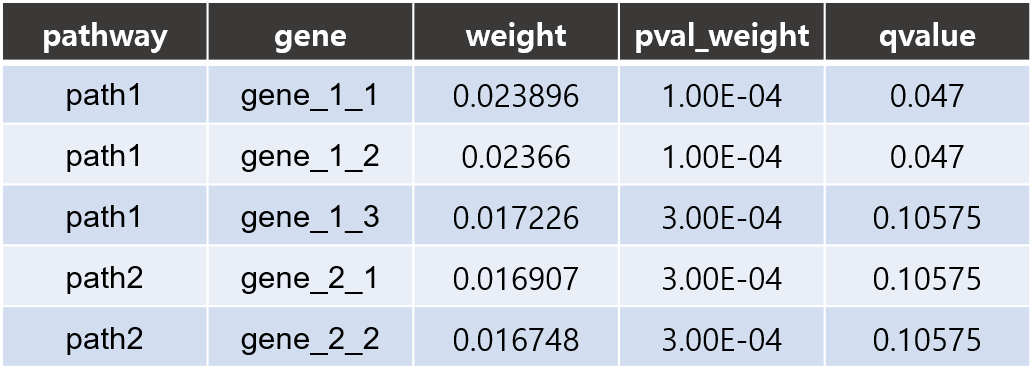
- Gene results I
- csv: Each line consists of 5 columns showing pathway name, gene name, gene coefficient(w_gene), p-value and FDR corrected q-value respectively.
The coefficient contained in this result file is about the weight (w_gene) value which represents the effect of the gene on the pathway.
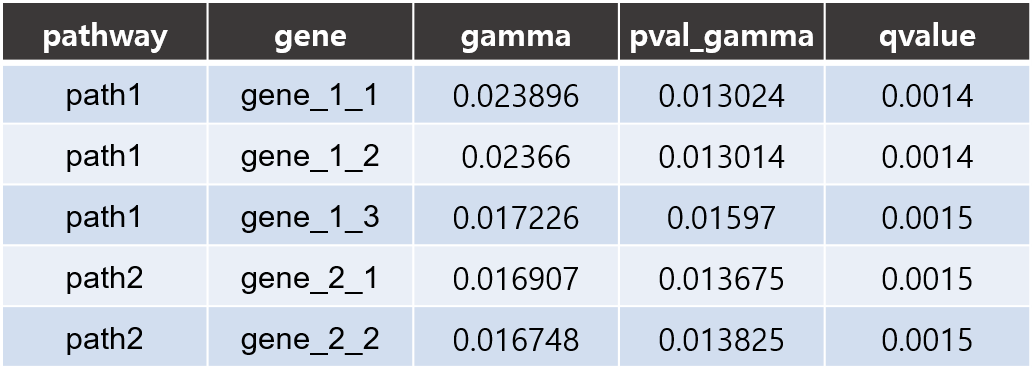
- Gene results II
- csv: Each line consists of 5 columns showing pathway name, gene name, gene coefficient(w_gene×β_path), p-value and FDR corrected q-value respectively.
The coefficients contained in this result file were calculated using the w_gene×β_path value for each gene which represents the effect of the gene on the phenotype.
Maintenance
If there is error, please let us know about the running problem to solve the problem and improve our program.
Contact : lydiamok25@snu.ac.kr